»The data analyst's three foundations in Python«
Matplotlib • Pandas • Jupyter Notebook
Matplotlib¶
- Standard for plotting with Python
- Recently v. 2.0.0 released
- → https://matplotlib.org/index.html
Using the global API¶
- Using the MATLAB-like interface
- Everything works through
plt.…
import matplotlib.pyplot as plt
x = range(10)
y = [i**2 for i in range(10)]
plt.plot(x, y)
plt.show()
Option Showcase¶
import numpy as np
x = np.arange(0, 100, 0.2)
y = np.sin(np.sqrt(x))
plt.plot(x, y, color="green")
plt.ylim([-0.6,1.1])
plt.xlabel("Numbers")
plt.ylabel("$\sin(\sqrt{Numbers})$")
plt.show()
Object API¶
- Instead of operation on global objects with
plt, rather useFigureandAxis(axes ≈ plots) - Cleaner approach (IMHO)
- Used under the hood of global API by leveraging
plt.gca().…(get current axis)
x = np.linspace(0, 2*np.pi, 400)
y = np.sin(x**2)
fig, ax = plt.subplots()
ax.plot(x, y)
ax.set_title('Use like this')
ax.set_xlabel("Numbers again")
<matplotlib.text.Text at 0x112c8fb38>
Multiple Plots¶
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, sharey=True)
ax1.plot(x, y)
ax1.set_title('Default Plot Style')
ax2.scatter(x, y, marker="D")
ax2.set_title('Scattered (Diamonds)')
fig.suptitle("Two Plots in One!")
<matplotlib.text.Text at 0x112dddf60>
Pandas¶
Introduction
Introduction¶
pandas is an open source, BSD-licensed library providing high-performance, easy-to-use data structures and data analysis tools for the Python programming language.
- Most important feature:
DataFrames and operations with them - → http://pandas.pydata.org/
import pandas as pd
Creating a DataFrame¶
Using a dictionary as an input
frame = pd.DataFrame({
"A": 1.2,
"B": pd.Timestamp('20170503'),
"C": [(-1)**i * np.sqrt(i) + np.e * (-1)**(i-1) for i in range(5)],
"D": pd.Categorical(["This", "column", "has", "entries", "entries"]),
"E": "Same"
})
frame
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 0 | 1.2 | 2017-05-03 | -2.718282 | This | Same |
| 1 | 1.2 | 2017-05-03 | 1.718282 | column | Same |
| 2 | 1.2 | 2017-05-03 | -1.304068 | has | Same |
| 3 | 1.2 | 2017-05-03 | 0.986231 | entries | Same |
| 4 | 1.2 | 2017-05-03 | -0.718282 | entries | Same |
Also available: .read_csv and .read_excel
Popular Functions on Frames¶
frame.describe()
| A | C | |
|---|---|---|
| count | 5.0 | 5.000000 |
| mean | 1.2 | -0.407224 |
| std | 0.0 | 1.781963 |
| min | 1.2 | -2.718282 |
| 25% | 1.2 | -1.304068 |
| 50% | 1.2 | -0.718282 |
| 75% | 1.2 | 0.986231 |
| max | 1.2 | 1.718282 |
frame.head(2)
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 0 | 1.2 | 2017-05-03 | -2.718282 | This | Same |
| 1 | 1.2 | 2017-05-03 | 1.718282 | column | Same |
Popular Functions on Frames II¶
frame.transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| A | 1.2 | 1.2 | 1.2 | 1.2 | 1.2 |
| B | 2017-05-03 00:00:00 | 2017-05-03 00:00:00 | 2017-05-03 00:00:00 | 2017-05-03 00:00:00 | 2017-05-03 00:00:00 |
| C | -2.71828 | 1.71828 | -1.30407 | 0.986231 | -0.718282 |
| D | This | column | has | entries | entries |
| E | Same | Same | Same | Same | Same |
frame.sort_values("C")
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 0 | 1.2 | 2017-05-03 | -2.718282 | This | Same |
| 2 | 1.2 | 2017-05-03 | -1.304068 | has | Same |
| 4 | 1.2 | 2017-05-03 | -0.718282 | entries | Same |
| 3 | 1.2 | 2017-05-03 | 0.986231 | entries | Same |
| 1 | 1.2 | 2017-05-03 | 1.718282 | column | Same |
Popular Functions on Frames III¶
round(frame,2)
frame.round(2)
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 0 | 1.2 | 2017-05-03 | -2.72 | This | Same |
| 1 | 1.2 | 2017-05-03 | 1.72 | column | Same |
| 2 | 1.2 | 2017-05-03 | -1.30 | has | Same |
| 3 | 1.2 | 2017-05-03 | 0.99 | entries | Same |
| 4 | 1.2 | 2017-05-03 | -0.72 | entries | Same |
frame.sum()
A 6.000000 C -2.036119 dtype: float64
frame.round(2).sum()
A 6.00 C -2.03 dtype: float64
Popular Functions on Frames IV¶
print(frame.round(2).to_latex())
\begin{tabular}{lrlrll}
\toprule
{} & A & B & C & D & E \\
\midrule
0 & 1.2 & 2017-05-03 & -2.72 & This & Same \\
1 & 1.2 & 2017-05-03 & 1.72 & column & Same \\
2 & 1.2 & 2017-05-03 & -1.30 & has & Same \\
3 & 1.2 & 2017-05-03 & 0.99 & entries & Same \\
4 & 1.2 & 2017-05-03 & -0.72 & entries & Same \\
\bottomrule
\end{tabular}
Index, Columns¶
frame["NewIdx"] = pd.date_range('20170504', periods=5)
frame.head(3)
| A | B | C | D | E | NewIdx | |
|---|---|---|---|---|---|---|
| 0 | 1.2 | 2017-05-03 | -2.718282 | This | Same | 2017-05-04 |
| 1 | 1.2 | 2017-05-03 | 1.718282 | column | Same | 2017-05-05 |
| 2 | 1.2 | 2017-05-03 | -1.304068 | has | Same | 2017-05-06 |
Index, Columns II¶
frame = frame.set_index("NewIdx") # Also: inplace=True
frame.head(3)
| A | B | C | D | E | |
|---|---|---|---|---|---|
| NewIdx | |||||
| 2017-05-04 | 1.2 | 2017-05-03 | -2.718282 | This | Same |
| 2017-05-05 | 1.2 | 2017-05-03 | 1.718282 | column | Same |
| 2017-05-06 | 1.2 | 2017-05-03 | -1.304068 | has | Same |
frame.index
DatetimeIndex(['2017-05-04', '2017-05-05', '2017-05-06', '2017-05-07',
'2017-05-08'],
dtype='datetime64[ns]', name='NewIdx', freq=None)
frame.columns
Index(['A', 'B', 'C', 'D', 'E'], dtype='object')
Slicing¶
Select only column "A"
frame["A"]
NewIdx 2017-05-04 1.2 2017-05-05 1.2 2017-05-06 1.2 2017-05-07 1.2 2017-05-08 1.2 Name: A, dtype: float64
Select columns "A" and "C"
frame[["A", "C"]].sort_values("C")
| A | C | |
|---|---|---|
| NewIdx | ||
| 2017-05-04 | 1.2 | -2.718282 |
| 2017-05-06 | 1.2 | -1.304068 |
| 2017-05-08 | 1.2 | -0.718282 |
| 2017-05-07 | 1.2 | 0.986231 |
| 2017-05-05 | 1.2 | 1.718282 |
Slicing II¶
frame[1:3]
| A | B | C | D | E | |
|---|---|---|---|---|---|
| NewIdx | |||||
| 2017-05-05 | 1.2 | 2017-05-03 | 1.718282 | column | Same |
| 2017-05-06 | 1.2 | 2017-05-03 | -1.304068 | has | Same |
frame.loc["2017-05-06"]
A 1.2 B 2017-05-03 00:00:00 C -1.30407 D has E Same Name: 2017-05-06 00:00:00, dtype: object
frame.iloc[2]
A 1.2 B 2017-05-03 00:00:00 C -1.30407 D has E Same Name: 2017-05-06 00:00:00, dtype: object
Slicing III¶
frame[frame["C"] > 0]
| A | B | C | D | E | |
|---|---|---|---|---|---|
| NewIdx | |||||
| 2017-05-05 | 1.2 | 2017-05-03 | 1.718282 | column | Same |
| 2017-05-07 | 1.2 | 2017-05-03 | 0.986231 | entries | Same |
frame[(frame["C"] > 0) & (frame["D"] == "has")]
| A | B | C | D | E | |
|---|---|---|---|---|---|
| NewIdx |
Plotting¶
frame[["A", "C"]].head(3)
| A | C | |
|---|---|---|
| NewIdx | ||
| 2017-05-04 | 1.2 | -2.718282 |
| 2017-05-05 | 1.2 | 1.718282 |
| 2017-05-06 | 1.2 | -1.304068 |
frame[["A", "C"]].plot()
<matplotlib.axes._subplots.AxesSubplot at 0x114187160>
Plotting II¶
frame[["A", "C"]].plot(
color=["red", "green"],
style=[".--","*"],
grid=True,
secondary_y=["C"]
)
<matplotlib.axes._subplots.AxesSubplot at 0x1141c75c0>
Plotting III¶
frame[["A", "C"]].plot(kind="bar")
<matplotlib.axes._subplots.AxesSubplot at 0x11433d5f8>
Plotting III (2)¶
frame[["A", "C"]].plot(kind="bar", stacked=True)
<matplotlib.axes._subplots.AxesSubplot at 0x1143d89b0>
Plotting III (3)¶
frame[["A", "C"]].reset_index().plot(kind="bar", subplots=True, figsize=(6,2))
array([<matplotlib.axes._subplots.AxesSubplot object at 0x1144b3438>,
<matplotlib.axes._subplots.AxesSubplot object at 0x114593668>], dtype=object)
Further
kinds:barh,box,hist,kde(a better histogram!),scatter; more: http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.plot.htmlInstead of
.plot(kind="bar"), also possible:.plot.bar()
Advanced Plotting¶
Combine Pandas & Matplotlib¶
Combine Pandas and Matplotlib by letting Pandas draw to an axis with ax
fig, ax = plt.subplots()
frame[["A", "C"]].plot(kind="bar", ax=ax)
ax.set_xlabel("Datetime")
ax.set_ylabel("Value")
fig.savefig("barplot.pdf")
Combination II¶
fig, (ax1, ax2, ax3) = plt.subplots(ncols=3, nrows=1, figsize=(12,3))
ax1 = frame["A"].plot.line(ax=ax1)
ax2 = frame["C"].plot.box(ax=ax2)
ax3 = frame["C"].plot.hist(ax=ax3, color="orange")
fig.suptitle("Stupid plots")
<matplotlib.text.Text at 0x1148029b0>
Seaborn¶
Seaborn is a library for making attractive and informative statistical graphics in Python
- Provides plotting interfaces
- And sets nice defaults
- Also: Colormaps
- → http://seaborn.pydata.org/
import seaborn as sns
sns.set(rc={"figure.figsize": (5, 3)})
frame["C"].plot(marker="s", linestyle="--")
<matplotlib.axes._subplots.AxesSubplot at 0x117fae240>
Seaborn Color Palette¶
frame["G"] = [(-1)**i * np.sqrt(i) + np.pi * (-1)**(i-1) for i in range(len(frame.index))]
frame["H"] = [(-1)**i * np.sqrt(i) + np.pi * (-1.1)**(i-1) for i in range(len(frame.index))]
with sns.color_palette("hls", 2):
fig, ax = plt.subplots()
sns.regplot(x="C", y="G", data=frame, ax=ax)
sns.regplot(x="C", y="H", data=frame, ax=ax)
Seaborn Color Palette II¶
sns.palplot(sns.color_palette())
sns.palplot(sns.color_palette("hls", 10))
sns.palplot(sns.color_palette("hls", 20))
sns.palplot(sns.color_palette("Paired", 10))
Seaborn Color Palette III / KDE Plot¶
x, y = np.random.multivariate_normal([0, 0], [[1, -.5], [-.5, 1]], size=300).T
cmap = sns.cubehelix_palette(light=1, as_cmap=True)
sns.kdeplot(x, y, cmap=cmap, shade=True);
Seaborn Color Palette IV / Jointplot¶
sns.jointplot(x=x, y=y, kind="reg")
<seaborn.axisgrid.JointGrid at 0x1188c15f8>
Complex Data¶
Some real data…¶
Some PAPI counters for different number of particles (=program run lengths), compiled with different compilers
dfCounters = pd.read_csv("juron-jube-add_one_to_list.csv")
dfCounters.head(2)
| modules | compiler | n_particles | hwc | HWC | |
|---|---|---|---|---|---|
| 0 | gcc/5.4.0 openmpi/2.0.2-gcc_5.4.0 PAPI/5.5.0-cuda | gfortran | 100000 | PAPI_TOT_INS | 32809671 |
| 1 | gcc/5.4.0 openmpi/2.0.2-gcc_5.4.0 PAPI/5.5.0-cuda | gfortran | 100000 | PAPI_TOT_CYC | 21246423 |
dfCounters = dfCounters.rename(columns={
"modules": "Modules",
"compiler": "Compiler",
"n_particles": "Number of Particles",
"hwc": "Counter Name",
"HWC": "Counter Value"
})
dfCounters.head(2)
| Modules | Compiler | Number of Particles | Counter Name | Counter Value | |
|---|---|---|---|---|---|
| 0 | gcc/5.4.0 openmpi/2.0.2-gcc_5.4.0 PAPI/5.5.0-cuda | gfortran | 100000 | PAPI_TOT_INS | 32809671 |
| 1 | gcc/5.4.0 openmpi/2.0.2-gcc_5.4.0 PAPI/5.5.0-cuda | gfortran | 100000 | PAPI_TOT_CYC | 21246423 |
Massaging¶
I want some relative values…
dfCounters["Counter Value (rel.)"] = dfCounters["Counter Value"] / dfCounters["Number of Particles"]
dfCounters.head(2)
| Modules | Compiler | Number of Particles | Counter Name | Counter Value | Counter Value (rel.) | |
|---|---|---|---|---|---|---|
| 0 | gcc/5.4.0 openmpi/2.0.2-gcc_5.4.0 PAPI/5.5.0-cuda | gfortran | 100000 | PAPI_TOT_INS | 32809671 | 328.09671 |
| 1 | gcc/5.4.0 openmpi/2.0.2-gcc_5.4.0 PAPI/5.5.0-cuda | gfortran | 100000 | PAPI_TOT_CYC | 21246423 | 212.46423 |
Some Values¶
Plot relative values of PAPI_TOT_CYC for gfortoran
dfCounters[
(dfCounters["Compiler"] == "gfortran")
&
(dfCounters["Counter Name"] == "PAPI_TOT_CYC")
]["Counter Value (rel.)"]\
.plot(marker="P")
<matplotlib.axes._subplots.AxesSubplot at 0x1185e66d8>
More Values¶
Plot same relativ values, but also those of counter PAPI_TOT_INS
dfCounters[
(dfCounters["Compiler"] == "gfortran")
&
((dfCounters["Counter Name"] == "PAPI_TOT_CYC") | (dfCounters["Counter Name"] == "PAPI_TOT_INS"))
]["Counter Value (rel.)"]\
.plot(marker="P")
<matplotlib.axes._subplots.AxesSubplot at 0x1186db4a8>
dfCounters[
(dfCounters["Compiler"] == "gfortran")
&
((dfCounters["Counter Name"] == "PAPI_TOT_CYC") | (dfCounters["Counter Name"] == "PAPI_TOT_INS"))
].head(3)
| Modules | Compiler | Number of Particles | Counter Name | Counter Value | Counter Value (rel.) | |
|---|---|---|---|---|---|---|
| 0 | gcc/5.4.0 openmpi/2.0.2-gcc_5.4.0 PAPI/5.5.0-cuda | gfortran | 100000 | PAPI_TOT_INS | 32809671 | 328.096710 |
| 1 | gcc/5.4.0 openmpi/2.0.2-gcc_5.4.0 PAPI/5.5.0-cuda | gfortran | 100000 | PAPI_TOT_CYC | 21246423 | 212.464230 |
| 5 | gcc/5.4.0 openmpi/2.0.2-gcc_5.4.0 PAPI/5.5.0-cuda | gfortran | 1000000 | PAPI_TOT_INS | 328081236 | 328.081236 |
fig, ax = plt.subplots()
ax = dfCounters[(dfCounters["Compiler"] == "gfortran") & (dfCounters["Counter Name"] == "PAPI_TOT_INS")]["Counter Value (rel.)"].plot(marker="P", ax=ax, label="PAPI_TOT_INS")
ax = dfCounters[(dfCounters["Compiler"] == "gfortran") & (dfCounters["Counter Name"] == "PAPI_TOT_CYC")]["Counter Value (rel.)"].plot(marker="o", ax=ax, label="PAPI_TOT_CYC")
ax.legend(loc="best", frameon=True, fontsize=15, framealpha=0.5)
ax.set_xlabel("Measurement number")
ax.set_ylabel("Counter Value (rel.)")
<matplotlib.text.Text at 0x118817c50>
Wouldn't be cool if Pandas could do this for us?
Pivoting!¶
Basically: Combine similar categorial data in a DataFrame
dfCounters.head(2)
| Modules | Compiler | Number of Particles | Counter Name | Counter Value | Counter Value (rel.) | |
|---|---|---|---|---|---|---|
| 0 | gcc/5.4.0 openmpi/2.0.2-gcc_5.4.0 PAPI/5.5.0-cuda | gfortran | 100000 | PAPI_TOT_INS | 32809671 | 328.09671 |
| 1 | gcc/5.4.0 openmpi/2.0.2-gcc_5.4.0 PAPI/5.5.0-cuda | gfortran | 100000 | PAPI_TOT_CYC | 21246423 | 212.46423 |
Some data massaging: I want to remove Modules column; but to prevent double-entries, I want to rename all mpifort Compiler entries run with module openmpi/1.10.2-pgi_16.10 loaded to PGI+MPI
dfCounters.loc[
dfCounters["Modules"].str.contains("openmpi/1.10.2-pgi_16.10")
&
(dfCounters["Compiler"] == "mpifort"),
"Compiler"
] = "PGI+MPI"
dfCounters = dfCounters.drop("Modules", axis=1)
dfCounters.head(2)
| Compiler | Number of Particles | Counter Name | Counter Value | Counter Value (rel.) | |
|---|---|---|---|---|---|
| 0 | gfortran | 100000 | PAPI_TOT_INS | 32809671 | 328.09671 |
| 1 | gfortran | 100000 | PAPI_TOT_CYC | 21246423 | 212.46423 |
Pivoting, Actually¶
index: What should be my new index? If array → hierarchical multi-indexvalues: What value should be printed in the cellscolumns: What should be the new columns? If array → hierarchical
dfPivot = dfCounters.pivot_table(
index="Number of Particles",
values="Counter Value (rel.)",
columns=["Compiler", "Counter Name"]
)
dfPivot.head(3)
| Compiler | PGI+MPI | gfortran | pgfortran | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Counter Name | PAPI_L1_DCM | PAPI_L2_DCM | PAPI_STL_ICY | PAPI_TOT_CYC | PAPI_TOT_INS | PAPI_L1_DCM | PAPI_L2_DCM | PAPI_STL_ICY | PAPI_TOT_CYC | PAPI_TOT_INS | PAPI_L1_DCM | PAPI_L2_DCM | PAPI_STL_ICY | PAPI_TOT_CYC | PAPI_TOT_INS |
| Number of Particles | |||||||||||||||
| 100000 | 3.032350 | 0.010760 | 479.309470 | 747.119030 | 780.156460 | 5.305490 | 0.002150 | 137.864120 | 212.464230 | 328.096710 | 1.088945 | 0.006175 | 232.514715 | 436.386840 | 672.144140 |
| 1000000 | 3.039885 | 0.008920 | 479.860810 | 747.309137 | 780.122863 | 2.744860 | 0.001581 | 137.640959 | 212.281606 | 328.081236 | 5.081417 | 0.005163 | 233.430218 | 436.841017 | 672.114449 |
| 2500000 | 3.419826 | 0.008527 | 479.873831 | 746.905123 | 780.120623 | 6.243841 | 0.001350 | 137.736993 | 214.782047 | 328.080074 | 9.485501 | 0.005273 | 233.018239 | 436.255372 | 672.112393 |
Pivot and Stack¶
Maybe getting the counters to the index side is more useful?
dfPivot.stack().head(6)
| Compiler | PGI+MPI | gfortran | pgfortran | |
|---|---|---|---|---|
| Number of Particles | Counter Name | |||
| 100000 | PAPI_L1_DCM | 3.032350 | 5.30549 | 1.088945 |
| PAPI_L2_DCM | 0.010760 | 0.00215 | 0.006175 | |
| PAPI_STL_ICY | 479.309470 | 137.86412 | 232.514715 | |
| PAPI_TOT_CYC | 747.119030 | 212.46423 | 436.386840 | |
| PAPI_TOT_INS | 780.156460 | 328.09671 | 672.144140 | |
| 1000000 | PAPI_L1_DCM | 3.039885 | 2.74486 | 5.081417 |
… which is the same as
dfCounters.pivot_table(
index=["Number of Particles", "Counter Name"],
values="Counter Value (rel.)",
columns="Compiler"
).head(6)
| Compiler | PGI+MPI | gfortran | pgfortran | |
|---|---|---|---|---|
| Number of Particles | Counter Name | |||
| 100000 | PAPI_L1_DCM | 3.032350 | 5.30549 | 1.088945 |
| PAPI_L2_DCM | 0.010760 | 0.00215 | 0.006175 | |
| PAPI_STL_ICY | 479.309470 | 137.86412 | 232.514715 | |
| PAPI_TOT_CYC | 747.119030 | 212.46423 | 436.386840 | |
| PAPI_TOT_INS | 780.156460 | 328.09671 | 672.144140 | |
| 1000000 | PAPI_L1_DCM | 3.039885 | 2.74486 | 5.081417 |
Plotting Pivoted DataFrames¶
dfPivot.plot(kind="bar", figsize=(12,5))
<matplotlib.axes._subplots.AxesSubplot at 0x1188ff2b0>
Plotting Pivoted DataFrames II¶
dfPivot.stack().plot(kind="bar", figsize=(11,5))
<matplotlib.axes._subplots.AxesSubplot at 0x118bedd68>
Plotting Pivoted DataFrames III¶
Focus on four counters, plot them next to each other
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(ncols=2, nrows=2, sharex=True, figsize=(12,5))
for (ax, counter) in zip([ax1, ax2, ax3, ax4], ["PAPI_TOT_INS", "PAPI_TOT_CYC", "PAPI_L1_DCM", "PAPI_STL_ICY"]):
ax = dfPivot.stack().loc[(slice(None), counter),:].plot(kind="bar", ax=ax, legend=False)
labels = [int(label.get_text().split(",")[0][1:-1]) for label in ax.get_xticklabels()]
ax.set_title(counter)
ax.set_xlabel("Number of Particles")
ax.set_ylabel("Counter Value per Particle")
ax.set_xticklabels(labels)
Jupyter Notebooks¶
Inline Magic I¶
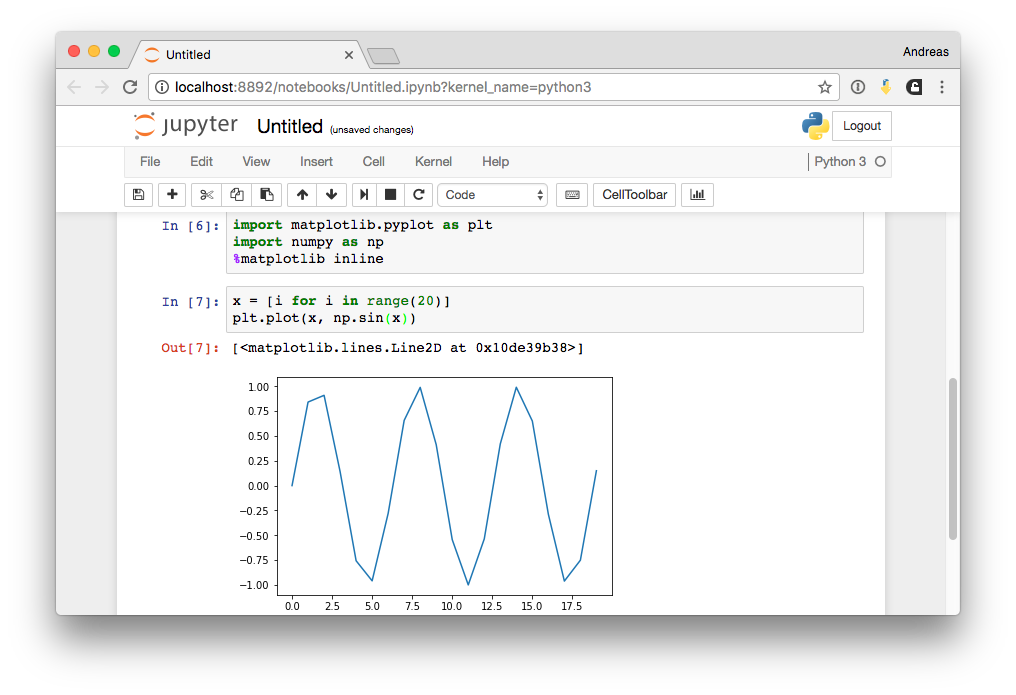
Inline Magic II¶
%timeit np.sin(range(1000))
1000 loops, best of 3: 698 µs per loop
%ls .
Pandas-Analysis.ipynb juron-jube-add_one_to_list.csv Pandas-Analysis.slides.html notebook-screenshot--inline1.png convertNotebookToHtmlSlides.sh* notebook-screenshot.png convertNotebookToPdfDocument.sh* reveal.js/ convertSlidesToPdf.sh* serveSlidesForPresentation.sh* custom.css
!pip install something
Collecting something Could not find a version that satisfies the requirement something (from versions: ) No matching distribution found for something
%lsmagic
Available line magics: %alias %alias_magic %autocall %automagic %autosave %bookmark %cat %cd %clear %colors %config %connect_info %cp %debug %dhist %dirs %doctest_mode %ed %edit %env %gui %hist %history %killbgscripts %ldir %less %lf %lk %ll %load %load_ext %loadpy %logoff %logon %logstart %logstate %logstop %ls %lsmagic %lx %macro %magic %man %matplotlib %mkdir %more %mv %notebook %page %pastebin %pdb %pdef %pdoc %pfile %pinfo %pinfo2 %popd %pprint %precision %profile %prun %psearch %psource %pushd %pwd %pycat %pylab %qtconsole %quickref %recall %rehashx %reload_ext %rep %rerun %reset %reset_selective %rm %rmdir %run %save %sc %set_env %store %sx %system %tb %time %timeit %unalias %unload_ext %who %who_ls %whos %xdel %xmode Available cell magics: %%! %%HTML %%SVG %%bash %%capture %%debug %%file %%html %%javascript %%js %%latex %%perl %%prun %%pypy %%python %%python2 %%python3 %%ruby %%script %%sh %%svg %%sx %%system %%time %%timeit %%writefile Automagic is ON, % prefix IS NOT needed for line magics.
Converting¶
- Notebooks are rendered at Github and our Gitlab server
- Can be converted to static HTML
- Can be converted to PDF, Markdown, reST, Slides
This presentation is one large Jupyter Notebook